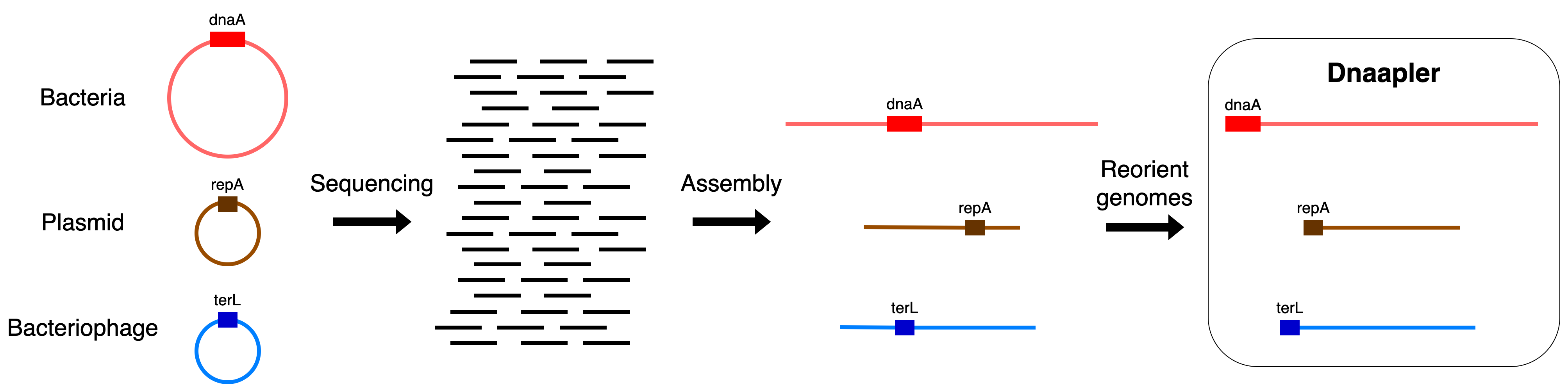
dnaapler is a simple python program that takes a single nucleotide input sequence (in FASTA format), finds the desired start gene using blastx against an amino acid sequence database, checks that the start codon of this gene is found, and if so, then reorients the chromosome to begin with this gene on the forward strand.
It was originally designed to replicate the reorientation functionality of Unicycler with dnaA, but for for long-read first assembled chromosomes. I have extended it to work with plasmids (dnaapler plasmid) and phages (dnaapler phage), or for any input FASTA desired with dnaapler custom,dnaapler largest, dnaapler mystery or dnaapler nearest.
If your input FASTA is mixed and you have 1 or more contigs (e.g. has chromosome and plasmids), you can also use dnaapler all, with the option to ignore some contigs with the --ignore parameter. This is probably the most useful command for most users.
Additionally, you can also reorient multiple bacterial chromosomes/plasmids/phages at once using the dnaapler bulk subcommand - it will give you more information about what contigs couldn't be rotated which may be useful.
For bacterial chromosomes, dnaapler chromosome should ensure the chromosome breakpoint never interrupts genes or mobile genetic elements like prophages. It is intended to be used with good-quality completed bacterial genomes, generated with methods such as Trycycler, Dragonflye or hybracter.
A clear schematic of what dnaapler is trying to achieve is presented below: